Workflow
This release note accompanies the R code used in the Practical session #2 - Mapping deep-water fishing grounds: from fishing tracks to fishing effort maps. It aims to detail part of the workflow adopted to elaborate Automatic Identification System (AIS) data in the context of the GFCM initiative to map fishing grounds for deep-water red shrimp in the Eastern-Central Mediterranean Sea. The processing workflow (Figure 1) is divided into three scripts and serve to (i) pre-process fishing tracks and to aggregate data for different purposes: (ii) fishing effort patterns and (iii) fishing pressure indicators. Results of points (ii) and (iii) are both visualized as map and as monthly time-series. Each step is shown as code chunk and it is briefly described.
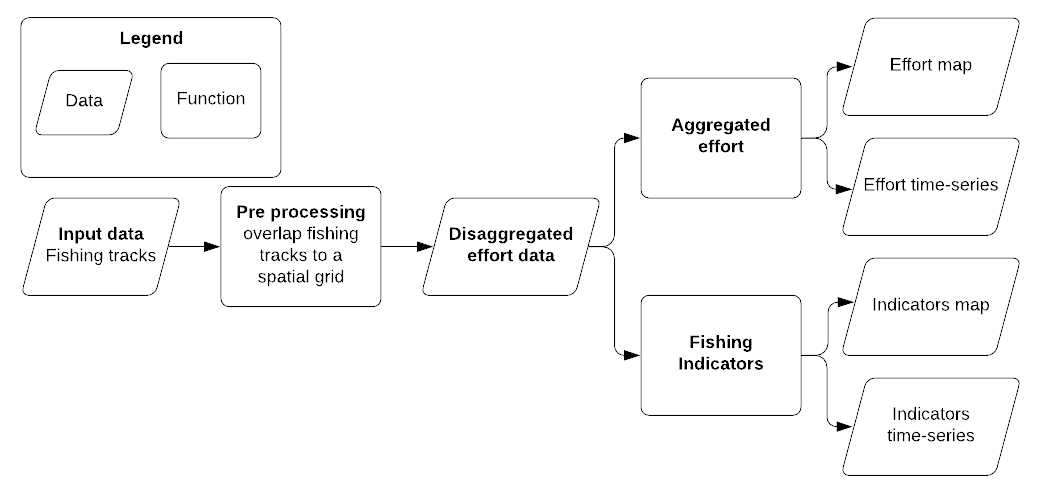
Figure 1: Conceptual workflow used in the Practical session #2 - Mapping deep-water fishing grounds: from fishing tracks to fishing effort maps
The results are some of the plots shown in the chapter 3.4 of the DWRS report on the fishing ground mapping (not yet published), in particular those referring to the aggregated fishing effort (Figure 2 a and b) and to the indicators of fishing pressure (Figure 2 c and d).
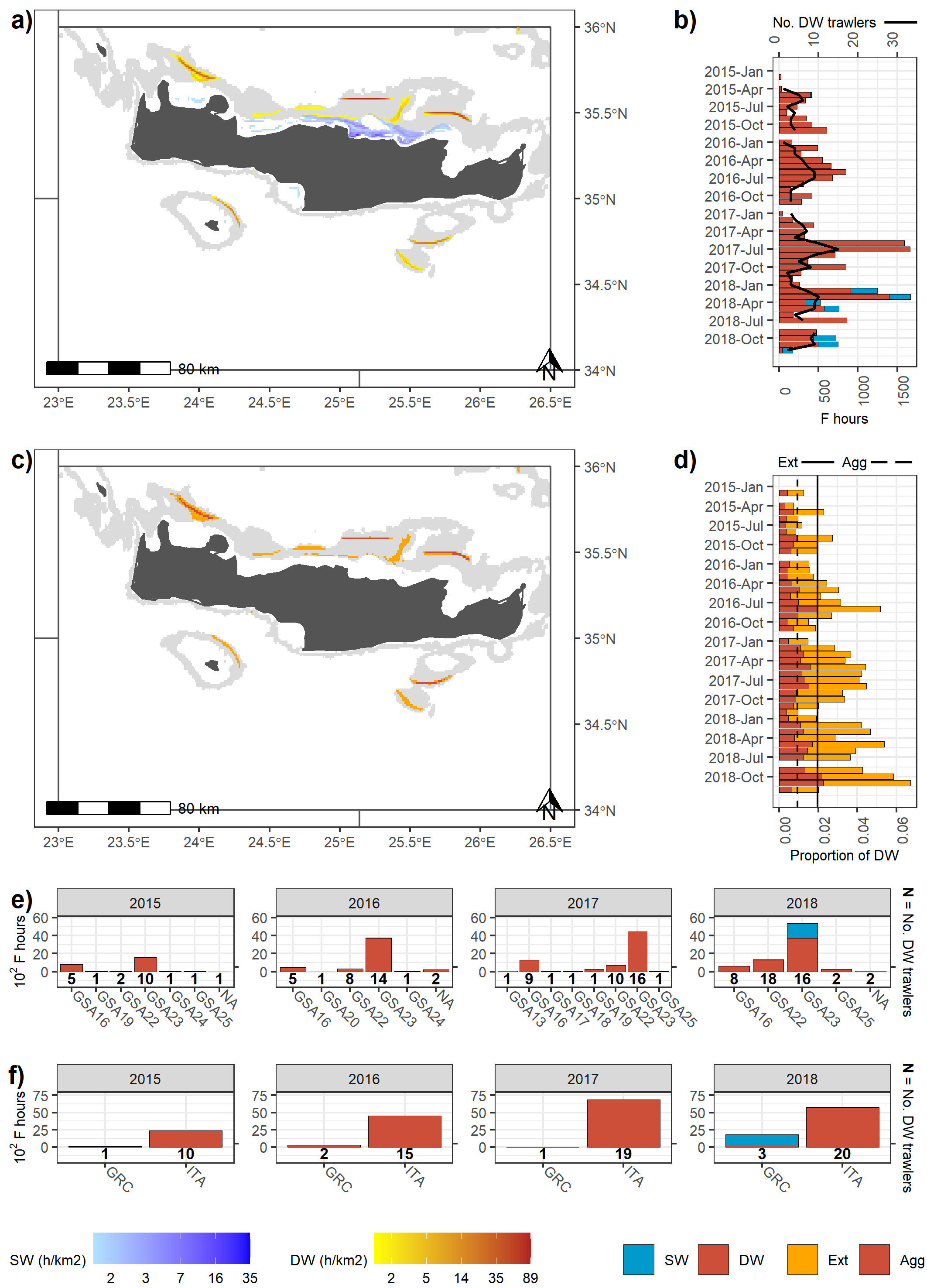
Figure 2: Fishing activities of vessels DW trawling in GSA 23. (a) Spatial distribution of mean DW and SW fishing activities during the study period; (b) Monthly fishing hours in DW (red) and SW (blue), and number of DW trawlers exerting these activities (solid line); (c) Spatial distribution of fishing pressure indicators of Extension (orange) and Aggregation (red) during the study period; (d) Monthly proportions of the DW area trawled and average values for fishing pressure indicators of extension (orange bars and solid line, respectively) and aggregation (red bars and dashed line, respectively); (e - f) Annual fishing hours in DW (red) and SW (blue) by GSA of arrival and country, and number of DW trawlers exerting these activities
Input data
Input data required to start the workflow are described below
Fishing data
The fishing data are loaded as f_tracks in the first script (see below) and are here briefly introduced:
| MMSI | year | month | gear | trip | id_track | s_time | f_time | range | geometry |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2015 | 2 | OTB | 126 | 85 | 2015-02-09 08:58:42 | 2015-02-09 10:43:52 | NA | LINESTRING (24.6528 35.0631… |
| 1 | 2015 | 2 | OTB | 126 | 86 | 2015-02-09 11:13:55 | 2015-02-09 13:19:10 | NA | LINESTRING (24.702 35.0109,… |
| 1 | 2015 | 2 | OTB | 127 | 87 | 2015-02-11 09:07:40 | 2015-02-11 09:42:43 | NA | LINESTRING (24.6769 35.0626… |
| 1 | 2015 | 2 | OTB | 127 | 88 | 2015-02-11 10:22:46 | 2015-02-11 13:53:04 | NA | LINESTRING (24.711 35.0207,… |
| 1 | 2015 | 2 | OTB | 127 | 89 | 2015-02-11 14:23:07 | 2015-02-11 14:43:07 | NA | LINESTRING (24.7117 35.0399… |
| 1 | 2015 | 2 | OTB | 127 | 90 | 2015-02-11 15:28:10 | 2015-02-11 16:39:43 | NA | LINESTRING (24.704 35.0097,… |
Where:
- MMSI: identifier of the vessel responsible for the fishing activity. Dummy data provided for privacy purposes.
- year: year in which the fishing activity was observed
- month: month in which the fishing activity was observed
- gear: gear predicted for the fishing activity
- trip: identifier of the trip where the fishing activity was observed
- id_track: identifier of the fishing track
- s_time: start time of the fishing track
- f_time: end time of the fishing track
- range: parameter that applies to Purse Seine fishing activity
- geometry: spatial attribute
Fishing data provided are distributed as in the map here shown:
Spatial layers
Fours vector layers are required to perform the functions foreseen by the workflow:
- grid: a 1km by 1km grid covering the GSA 23, with additional attributes indicating the bottom type and if the cell is or not within the 400-800 m bathymetry.
- land: contour of land
- gsa: boundaries of the GFCM-GSA
- dep: contour of the 400-800m batymetry
grid <- read_sf("maps/grid")
land=read_sf("maps/land")%>%
st_crop(., xmin=-10, xmax=43, ymin=20, ymax=48) #
gsa<-read_sf("maps/gsa")%>%
dplyr::filter(SMU_CODE == '23' )
dep = read_sf("maps/depth_contours")%>%
st_cast(., to = "POLYGON")%>%
st_set_crs(st_crs(gsa))%>%
st_buffer(0)%>%
st_crop(gsa)
lims=c(st_bbox(gsa[c(1,3)], st_bbox(gsa[c(2,4)])))
Processing
The proposed workflow is hereafter applied on the fishing tracks of a single area although it can be iterated on multiple areas. It is organized into three scripts:
- PreProcessing: it aims to overlap fishing tracks to a spatial grid.
- FishingPattern: it starts from the output of the first script and it aggregate data to produce and visualize fishing effort pattern
- FishingIndicators: it starts from the output of the first script and it aggregate data to produce and visualize fishing effort indicators
Pre processing
The topic of this first section is the overlap of the fishing track to a grid covering the reference area. This is a key-step and its output will represent the basis for all the images contained in the Figure 2.
Before to start
Check the working directory (WD), which have to be “practical_session_2”. The getwd() command shows you which is your WD. If you just opened the project, push on the setwd('practical_session_2/') line. In case you need to go back of one folder, the setwd('..') command sets the WD to a backward place.
Load spatial Data and set parameters
The first part of the code aims to prepare the environment for the processing of the fishing data. Loading the spatial grid:
grid=read_sf("maps/grid")Defining two parameters:
activate_filteractivates a filter to the data to remove outliers (see below in Step 4)ref_yearsindicate the time span of the analysis
activate_filter='Y'
ref_years=2015:2018For loop sectioned
In the original code, all the following steps are done within a for loop iterating on the years (as defined by ref_years). For illustrative purposes, here we present all the operations outside the for loop, taking the year 2015 as example.
Step 1
Load the fishing data for one year (2015 used as example)
f_tracks=read_sf(file.path("data",2015,paste0("f_segments_DWRS_WS_", 2015, ".shp")))
f_tracks$s_time=as_datetime(f_tracks$s_time, tz='UTC')
f_tracks$f_time=as_datetime(f_tracks$f_time, tz='UTC')Step 2
The next step deals with applying the estimate_fishing_effort_DWRS function on the data for each vessel. This function, which is inherited from the R4ais workflow, is defined in a separate script. Here below is shown the original code. The function performs a spatial intersection between the fishing tracks and the grid cells, then it aggregates the results to obtain the fishing activity occurring within each grid cell. The difference with the R4ais version is in the factors used to aggregate the data: data are aggregated by the factors grid_id+gear+MMSI+trip+s_time, while in the R4ais only by grid_id+gear. This modification was needed to keep data at the maximum level of disaggregation, permitting later to use the same dataset for multiple purposes.
estimate_fishing_effort_DWRS <- function(fishing_tracks, grid){
lapply(fishing_tracks, function(x){
if(is.data.frame(x)){
x = st_sf(x)
}
xgear = unique(x$gear)
st_crs(x)=wgs # set crs
if(xgear == "PS"){
x$duration=difftime(x$f_time, x$s_time, units="secs")
x=st_as_sf(x)
xint=st_intersection(grid, x)
xint=data.frame(xint)
xint=xint[,c("grid_id", "gear", "duration")]
xint$fishing_hours=xint$duration/3600
f_hours = aggregate(fishing_hours~grid_id+gear, data = xint, sum)
colnames(f_hours)[ncol(f_hours)] = "f_hours"
grid_edit=merge(grid, f_hours, by="grid_id") # combine effort to grid
return(grid_edit)
}else{
xnames=names(x)
x$distance=st_length(x$geometry, units="m") # estimated fishing track
x$duration=difftime(x$f_time, x$s_time, units="secs")
x$speed_ms=as.numeric(x$distance)/as.numeric(x$duration)
x$speed_kn=x$speed_ms*1.94384
x=st_as_sf(x)
xint=st_intersection(grid, x)
if(nrow(xint) > 0){
xint$observed_swept=st_length(xint)
xint=data.frame(xint)
xint=xint[,c("grid_id","MMSI", "trip", "s_time" ,"gear","observed_swept", "speed_ms")]
xint$fishing_hours=(as.numeric(xint$observed_swept)/xint$speed_ms)/3600
xint
f_hours = aggregate(fishing_hours~grid_id+gear+MMSI+trip+s_time, data = xint, sum) # estimate fishing effort in cells grid
colnames(f_hours)[ncol(f_hours)] = "f_hours"
grid_edit=merge(grid, f_hours, by="grid_id") # combine effort to grid
grid_edit=grid_edit[grid_edit$f_hours>0,]
grid_edit=grid_edit[,c("MMSI","gear", "trip", "s_time" , 'G_ID', 'DW', 'f_hours')]
return(grid_edit)
}
}
})
}Data are first divided into a list by vessel identifier and then the estimate_fishing_effort_DWRS function is applied on each element of the list.
f_tracks=split(f_tracks, f_tracks$MMSI)
f_tracks_grid=estimate_fishing_effort_DWRS(f_tracks, grid)Step 3
As the output of the estimate_fishing_effort_DWRS function is a list, the data need to be unlisted and formatted. Later on, the information regarding year and month of the fishing activity it is added.
f_tracks_grid=plyr::ldply(f_tracks_grid,.id = NULL)
f_tracks_grid=f_tracks_grid[ , -which(names(f_tracks_grid) %in% c("grid_id", 'geometry'))] ## remove not useful information
f_tracks_grid$month=lubridate::month(f_tracks_grid$s_time)
f_tracks_grid$year=lubridate::year(f_tracks_grid$s_time)Step 4 (Optional)
To remove some outliers, we may exclude grid cells where fishing activities over the entire year were fished for less than 0.33 hours.
if(activate_filter=="Y"){
print("Filtering activated")
outlier_filter=aggregate(x=f_tracks_grid$f_hours, by=list('G_ID'=f_tracks_grid$G_ID,'year'=f_tracks_grid$year ), FUN=sum)
outlier_filter=outlier_filter[outlier_filter$x>=0.33,c('G_ID','year')]
f_tracks_grid=merge(outlier_filter, f_tracks_grid, by=c("G_ID", "year"), all.x=T)
}## [1] "Filtering activated"Results
The resulting data fishing_data is the base grid having the cells filled with the information of fishing activity. Each row refers to the intersection between a grid cell G_ID and a unique fishing track defined by its starting time s_time and the fishing vessel MMSI performing it. Therefore each fishing track is divided into as many rows as the number of grid cells intersected. This type of information permits to aggregate the fishing activity in all the format needed for the further analysis.
Here it is shown the header of the dataset.
| G_ID | year | MMSI | gear | trip | s_time | DW | f_hours | month |
|---|---|---|---|---|---|---|---|---|
| 1872289 | 2015 | 1 | OTB | 149 | 2015-09-25 18:58:07 | 1 | 0.2296348 | 9 |
| 1872289 | 2015 | 1 | OTB | 151 | 2015-09-29 06:35:27 | 1 | 0.2491466 | 9 |
| 1872289 | 2015 | 1 | OTB | 149 | 2015-09-25 08:32:07 | 1 | 0.1997793 | 9 |
| 1872289 | 2015 | 1 | OTB | 150 | 2015-09-28 16:49:15 | 1 | 0.2390655 | 9 |
| 1872289 | 2015 | 1 | OTB | 149 | 2015-09-25 09:37:10 | 1 | 0.0942475 | 9 |
| 1872290 | 2015 | 1 | OTB | 150 | 2015-09-28 16:49:15 | 1 | 0.2367738 | 9 |
Where:
- G_ID: id of the grid cell
- year: year in which the fishing activity was observed
- MMSI: identifier of the vessel responsible for the fishing activity
- gear: gear predicted for the fishing activity
- trip: identifier of the trip where the fishing activity was observed
- s_time: start time of the fishing track (not related to the grid cell)
- DW: binary column indicating if the grid cell falls in the DW stratum.
- f_hours: fishing hours observed in the
G_IDgrid cell for the specific fishing track starting ats_timeand performed by theMMSIvessel - month: month in which the fishing activity was observed
FishingPattern
This section presents the code used to produce the Figure 35.a of the DWS-Fishing activity report. It is a thematic map showing the mean fishing hours by grid cell in the GSA 23, over the period january 2015-december 2018.
Before to start
Check the working directory (WD), which have to be “practical_session_2”. The getwd() command shows you which is your WD. If you just opened the project, push on the setwd('practical_session_2/') line. In case you need to go back of one folder, the setwd('..') command sets the WD to a backward place.
Load data
The input data is represented by the output of the PreProcessing.R script and by the spatial data introduced at the beginning of the lesson. In addition, it is loaded an R script containing the default setting for the base map used.
fishing_data= read_csv("data/fishing_data.csv")
rm(vessel_selection)
# Shapefiles
grid <- read_sf("maps/grid")
land=read_sf("maps/land")%>%
st_crop(., xmin=-10, xmax=43, ymin=20, ymax=48)
gsa=read_sf("maps/gsa")%>%
dplyr::filter(SMU_CODE == '23' )
dep = read_sf("maps/depth_contours")%>%
st_cast(., to = "POLYGON")%>%
st_set_crs(st_crs(gsa))%>%
st_buffer(0)%>%
st_crop(gsa)
lims=c(st_bbox(gsa[c(1,3)], st_bbox(gsa[c(2,4)])))
text_size=8
plot_height=25
plot_width=18
source("R/supporting_code/plot_specifications.R")defo_map
Optional
In the following line there is the possibility to reduce the dataset by selecting only those vessels that fished in the DW strata at least once in a month.
vessel_selection=unique(fishing_data[fishing_data$DW==1,c('MMSI','month','year')]) # Create a monthly list of vessels fishing in the DW strata
fishing_data=merge(vessel_selection, fishing_data, by=c("MMSI", "year", "month"), all.x = T)Effort Map
Step 1
The fishing_data dataset is aggregated to get mean annual fishing intensity by grid cell. This is done in two consecutive aggregation functions. The cumulative_hours_by_year dataset is the annual cumulative values of fishing activity by grid cell. In the following line, the same dataset is further summarized into mean_hours_over_year, which is the mean value by cell over the years.
cumulative_hours_by_year=aggregate(list(f_hours_year=fishing_data$f_hours),
by=list('G_ID'= fishing_data$G_ID ,'year'=fishing_data$year), FUN=sum)
mean_hours_over_year=aggregate(list(f_hours_overall=cumulative_hours_by_year$f_hours_year),
by=list('G_ID'=cumulative_hours_by_year$G_ID),FUN=mean)
|
Step 2
Now that the overall fishing intensity by grid cell is obtained, it is possible to paste this information to the base grid. This is done by using the merge function, using the G_ID column as primary key. An additional column with the log of fishing hours is also created
grid_fishing=merge(grid, mean_hours_over_year, by='G_ID', all.x=F)
grid_fishing$f_hours_log=log(grid_fishing$f_hours + 1)| G_ID | DW | GSA | EUNIScomb | grid_id | f_hours_overall | geometry | f_hours_log |
|---|---|---|---|---|---|---|---|
| 1867864 | 1 | GSA23 | A6.511 | 15016 | 0.5300249 | POLYGON ((23.79001 35.84624… | 0.4252840 |
| 1868595 | 1 | GSA23 | A6.511 | 15186 | 0.3755053 | POLYGON ((23.80001 35.84624… | 0.3188211 |
| 1869328 | 1 | GSA23 | A6.511 | 15357 | 0.4563027 | POLYGON ((23.81001 35.84624… | 0.3759008 |
| 1870067 | 1 | GSA23 | A6.511 | 15529 | 0.4577012 | POLYGON ((23.82001 35.83624… | 0.3768607 |
| 1870808 | 1 | GSA23 | A6.511 | 15700 | 1.2239307 | POLYGON ((23.83001 35.83624… | 0.7992762 |
| 1870810 | 1 | GSA23 | A6.511 | 15702 | 0.4821836 | POLYGON ((23.83001 35.81624… | 0.3935164 |
This is how the grid populated with fishing activity appears. The interactive map is just for illustrative purposes and it is not part of the R code used for the course.
Step 3
To facilitate the plotting, grid data are divided by depth in two separate files (grid_DW and grid_SW). Fishing hours are also normalized.
grid_DW=grid_fishing[grid_fishing$DW==1,]
grid_DW$f_hours_log_st=st_fun(grid_DW$f_hours_log)
grid_SW=grid_fishing[grid_fishing$DW==0,]
grid_SW$f_hours_log_st=st_fun(grid_SW$f_hours_log)Plot
Specifications for the plot
Plots are standardized between figures using the settings loaded at the beginning of the script. In addition, labeling can be optimized to fit the data of each area.
brks_DW = seq(0, 1, length.out = 10)[c(2,4,6,8,10)]
brks_DW_lab = round(exp((brks_DW*(max(grid_DW$f_hours_log) - min(grid_DW$f_hours_log))) + min(grid_DW$f_hours_log)))
brks_SW = seq(0, 1, length.out = 10)[c(2,4,6,8,10)]
brks_SW_lab = round(exp((brks_SW*(max(grid_SW$f_hours_log) - min(grid_SW$f_hours_log))) + min(grid_SW$f_hours_log)))Result
The object defo_map contains all the settings specified above and represents the basis of our plot. Here are added the effort statistics, along with the parameters to set the legend values appropriately.
plot_effort_map = defo_map+
geom_sf(data = grid_SW, aes(fill = f_hours_log_st), colour = NA, alpha = 0.8, show.legend = "point" ) +
scale_fill_gradient(low = "lightskyblue1", high = "blue", breaks = brks_SW, labels = brks_SW_lab, guide = guide_colorbar(title = "SW (h/km2)")) +
new_scale("fill") +
geom_sf(data = grid_DW, aes(fill = f_hours_log_st), colour = NA) +
scale_fill_gradient("DW (h/km2)", breaks = brks_DW, labels = brks_DW_lab, low = "yellow", high = "firebrick") +
coord_sf(xlim=c(lims[[1]], lims[[3]]), ylim=c(lims[[2]], lims[[4]]) , label_axes = list(bottom = "E", right = "N")) +
xlab("") ; plot_effort_map## Coordinate system already present. Adding new coordinate system, which will replace the existing one.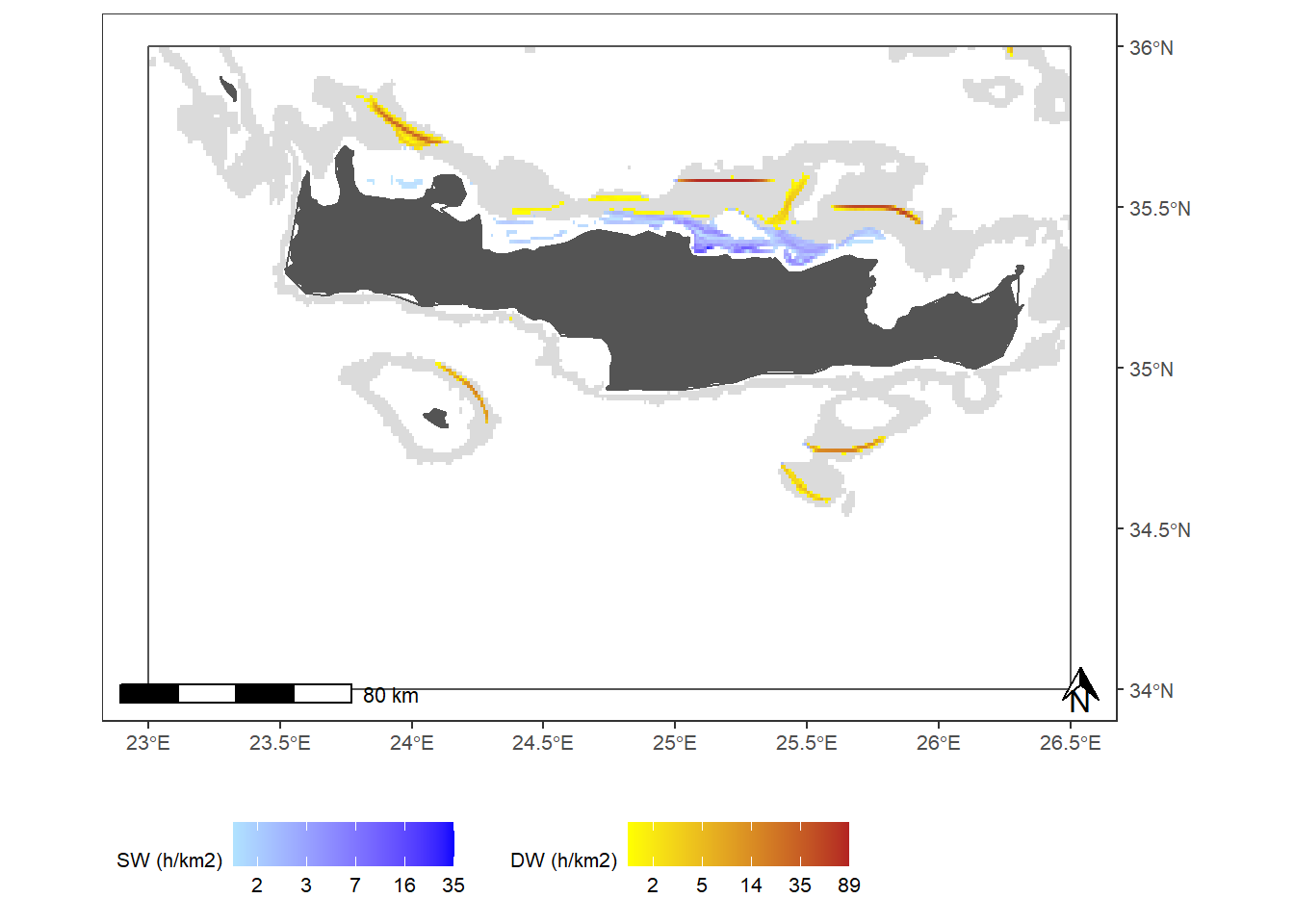
Fishing effort monthly time series
Step 1
The procedure is similar to what described in the Fishing effort map section, although here the values are calculate on monthly basis and summarized.
The monthly_effort dataset contains the cumulative monthly fishing hours by grid cell. In an additional line of code it is pasted a formatted information on the reference month.
monthly_effort=aggregate(list(nhours=fishing_data$f_hours),
by=list('year'= fishing_data$year, 'month'= fishing_data$month ,'DW'=fishing_data$DW), FUN=sum)
monthly_effort$temporal_ref=as.Date(paste(monthly_effort$year, monthly_effort$month, "01", sep = "-"))| year | month | DW | nhours | temporal_ref |
|---|---|---|---|---|
| 2016 | 1 | 0 | 0.6321288 | 2016-01-01 |
| 2018 | 1 | 0 | 0.0290744 | 2018-01-01 |
| 2017 | 2 | 0 | 0.0701169 | 2017-02-01 |
| 2018 | 2 | 0 | 344.5507970 | 2018-02-01 |
| 2016 | 3 | 0 | 0.3357668 | 2016-03-01 |
| 2017 | 3 | 0 | 0.5325699 | 2017-03-01 |
The vessel_by_month dataset contanins the monthly number of vessels fishing in the case study area, both for the DW and the SW batimetries. In an additional line of code it is pasted a formatted information on the reference month.
vessel_by_month=unique(fishing_data[,c('MMSI','month','year')])
vessel_by_month=aggregate(vessel_by_month$MMSI,
by=list('year'=vessel_by_month$year , 'month'=vessel_by_month$month), FUN=length)
vessel_by_month$temporal_ref=as.Date(paste(vessel_by_month$year, vessel_by_month$month, "01", sep = "-"))| year | month | x | temporal_ref |
|---|---|---|---|
| 2016 | 1 | 1 | 2016-01-01 |
| 2017 | 1 | 3 | 2017-01-01 |
| 2018 | 1 | 3 | 2018-01-01 |
| 2015 | 2 | 2 | 2015-02-01 |
| 2016 | 2 | 4 | 2016-02-01 |
| 2017 | 2 | 4 | 2017-02-01 |
Step 2
The monthly fishing hours and vessel statistics are calculated only for those months were fishing activity was actually observed. However we aim to consider in the plot also the months where no activity at all was observed. Therefore some formatting lines are needed.
index=data.frame(order=seq(1, 48, 1),
temporal_ref=seq(as.Date(paste("2015", "01", "01", sep = "-")),
as.Date(paste("2018", "12", "01", sep = "-")),"month"))
monthly_effort=merge(index, monthly_effort, by="temporal_ref", all.x = T)
monthly_effort[!is.na(monthly_effort$DW) & monthly_effort$DW==1,]$DW='DW'
monthly_effort[!is.na(monthly_effort$DW) & monthly_effort$DW==0,]$DW='SW'
vessel_by_month=merge(index, vessel_by_month, by="temporal_ref", all.x = T)Plot
Specifications for the plot
Plots are standardized between figures using the settings loaded at the beginning of the script. In addition, labeling can be optimized to fit the data of each area.
plot_breaks=seq(min(monthly_effort$order),max(monthly_effort$order),3)
plot_labels=paste(substr(seq(min(monthly_effort$temporal_ref),
max(monthly_effort$temporal_ref),"quarters"),1,4),
month.abb[as.numeric(substr(seq(min(monthly_effort$temporal_ref),max(monthly_effort$temporal_ref),"quarters"),6,7))], sep="-")Result
plot_effort_ts=defo_ts +
geom_bar(data = monthly_effort, aes(x=order,y= nhours, fill = reorder(DW, desc(DW))),
stat = "identity", colour = 1, size=0.2) +
scale_fill_manual(values = c("deepskyblue3", "tomato3")) +
xlab("") +
scale_x_reverse(breaks=plot_breaks,labels=plot_labels)+
guides(fill = guide_legend(title = "Fishing ground")) +
ylab("Hours") +
geom_line(data=vessel_by_month, aes(x=order, y = x*50), size = 0.7) +
scale_y_continuous(sec.axis = sec_axis(~ . /50, name=expression(paste(""[""["No. DW trawlers"]], paste(bold(" __"))))), name=expression( ""[""["F hours"]])) ; plot_effort_ts## Warning: Removed 5 rows containing missing values (position_stack).## Warning: Removed 1 row(s) containing missing values (geom_path).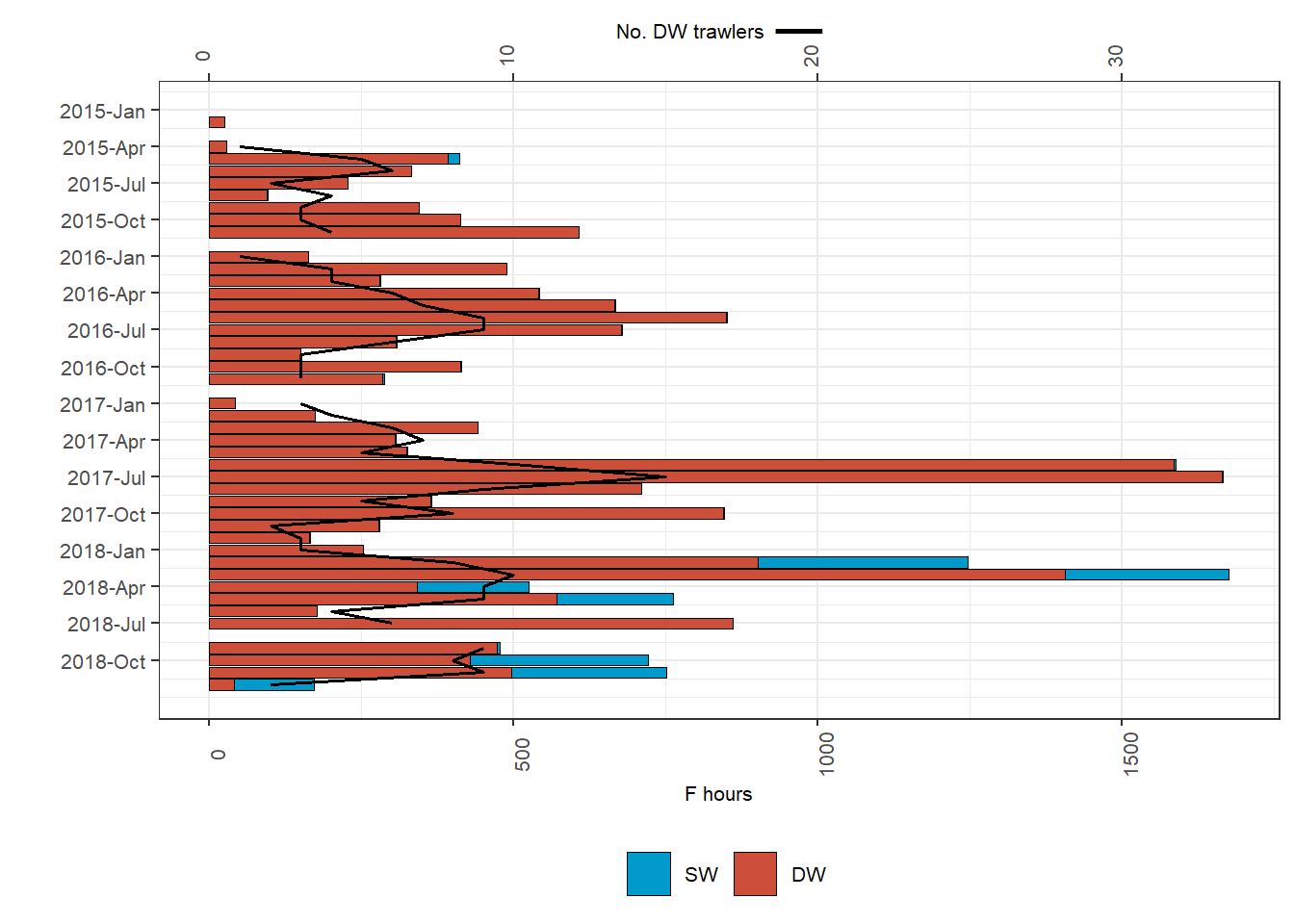
FishingIndicators
Before to start
Check the working directory (WD), which have to be “practical_session_2”. The getwd() command shows you which is your WD. If you just opened the project, push on the setwd('practical_session_2/') line. In case you need to go back of one folder, the setwd('..') command sets the WD to a backward place.
Load data
The input data is represented by the output of the PreProcessing.R script and by the spatial data introduced at the beginning of the lesson. In addition, it is loaded an R script containing the default setting for the base map used.
fishing_data= read_csv("data/fishing_data.csv")
rm(vessel_selection)
# Shapefiles
grid <- read_sf("maps/grid")
land=read_sf("maps/land")%>%
st_crop(., xmin=-10, xmax=43, ymin=20, ymax=48)
gsa=read_sf("maps/gsa")%>%
dplyr::filter(SMU_CODE == '23' )
dep = read_sf("maps/depth_contours")%>%
st_cast(., to = "POLYGON")%>%
st_set_crs(st_crs(gsa))%>%
st_buffer(0)%>%
st_crop(gsa)
lims=c(st_bbox(gsa[c(1,3)], st_bbox(gsa[c(2,4)])))
text_size=8
plot_height=25
plot_width=18
source("R/supporting_code/plot_specifications.R")Optional
In the following line there is the possibility to reduce the dataset by selecting only those vessels that fished in the DW strata at least once in a month.
vessel_selection=unique(fishing_data[fishing_data$DW==1,c('MMSI','month','year')]) # Create a monthly list of vessels fishing in the DW strata
fishing_data=merge(vessel_selection, fishing_data, by=c("MMSI", "year", "month"), all.x = T)Indicator Map
Step 1
The indicator map is performed only for the DW stratum and over the entire reference period. As so, the first step requires to filter the input data to the reference batymetric stratum and to aggregate the fishing activity by grid cell over the 2015-208 period.
DWfishing_data=fishing_data[fishing_data$DW==1,]
DWfishing_data=aggregate(list('f_hours'=DWfishing_data$f_hours),
by=list('G_ID'= DWfishing_data$G_ID), FUN=sum)| G_ID | f_hours |
|---|---|
| 1867864 | 0.5300249 |
| 1868595 | 0.3755053 |
| 1869328 | 0.9126055 |
| 1870067 | 0.9154025 |
| 1870808 | 2.4478613 |
| 1870810 | 0.9643671 |
Step 2
The successive step relates to the calculation of the indicators. The dedicated functions are stored in a separate file for tidiness purpose and are shown below. The ind5 function serve to calculate the MSFD indicator D5, namely the extension of the fishing activity in a given area. It calculates the number of cells of the grid where fishing activity was observed over the reference period.
ind5=function(x){
x = unique(x[,c('G_ID')])
return(x)
}The ind6 serve to calculate the MSFD indicator D6, namely the aggregation of the fishing activity in a given area. It calculates the number of cells of the grid containing the 90% of the cumulative distribution of the fishing activity over the refere
ind6=function(x){
x = x[order(x$f_hours, decreasing = T),]
x$cum = cumsum(x$f_hours)
xthr = sum(x$f_hours, na.rm = T) * 0.9
x=x[which(x$cum <= xthr),]
return(x)
}Both functions are applied to the data.
ind5_map=ind5(DWfishing_data)
ind6_map=ind6(DWfishing_data)
|
|
Step 3
Add the mean overall fishing intensity to the grid shapefile. For indicator 5 it is enough to filter the grid. For indicator 6 is done by using the merge function, using the G_ID column as primary key.
grid_ind5=grid[grid$G_ID%in%ind5_map,]
grid_ind6=merge(grid, ind6_map, by='G_ID', all.x=F)| G_ID | DW | GSA | EUNIScomb | grid_id | f_hours | cum | geometry |
|---|---|---|---|---|---|---|---|
| 1873032 | 1 | GSA23 | A6.511 | 16217 | 42.63207 | 17813.37 | POLYGON ((23.86001 35.81624… |
| 1873776 | 1 | GSA23 | A6.511 | 16389 | 62.85778 | 16564.71 | POLYGON ((23.87001 35.80624… |
| 1874518 | 1 | GSA23 | A6.511 | 16562 | 96.66239 | 15052.40 | POLYGON ((23.88001 35.79624… |
| 1875256 | 1 | GSA23 | A6.511 | 16734 | 38.96086 | 18056.29 | POLYGON ((23.89001 35.79624… |
| 1875257 | 1 | GSA23 | A6.511 | 16735 | 91.00845 | 15520.13 | POLYGON ((23.89001 35.78624… |
| 1875994 | 1 | GSA23 | A6.511 | 16907 | 87.69846 | 15787.08 | POLYGON ((23.90001 35.78624… |
Plot
Specifications for the plot
Plots are standardized between figures using the settings loaded at the beginning of the script.
Result
plot_indicator_map = defo_map+
geom_sf(data = grid_ind5, aes(fill = "orange"), colour = NA) +
geom_sf(data = grid_ind6, aes(fill = "tomato3"), colour = NA) +
scale_fill_identity(guide='legend',
breaks = c("orange", "tomato3"),
labels = c("Ext", "Agg"),
name='')+
coord_sf(xlim=c(lims[[1]], lims[[3]]), ylim=c(lims[[2]], lims[[4]]) , label_axes = list(bottom = "E", right = "N"));plot_indicator_map## Coordinate system already present. Adding new coordinate system, which will replace the existing one.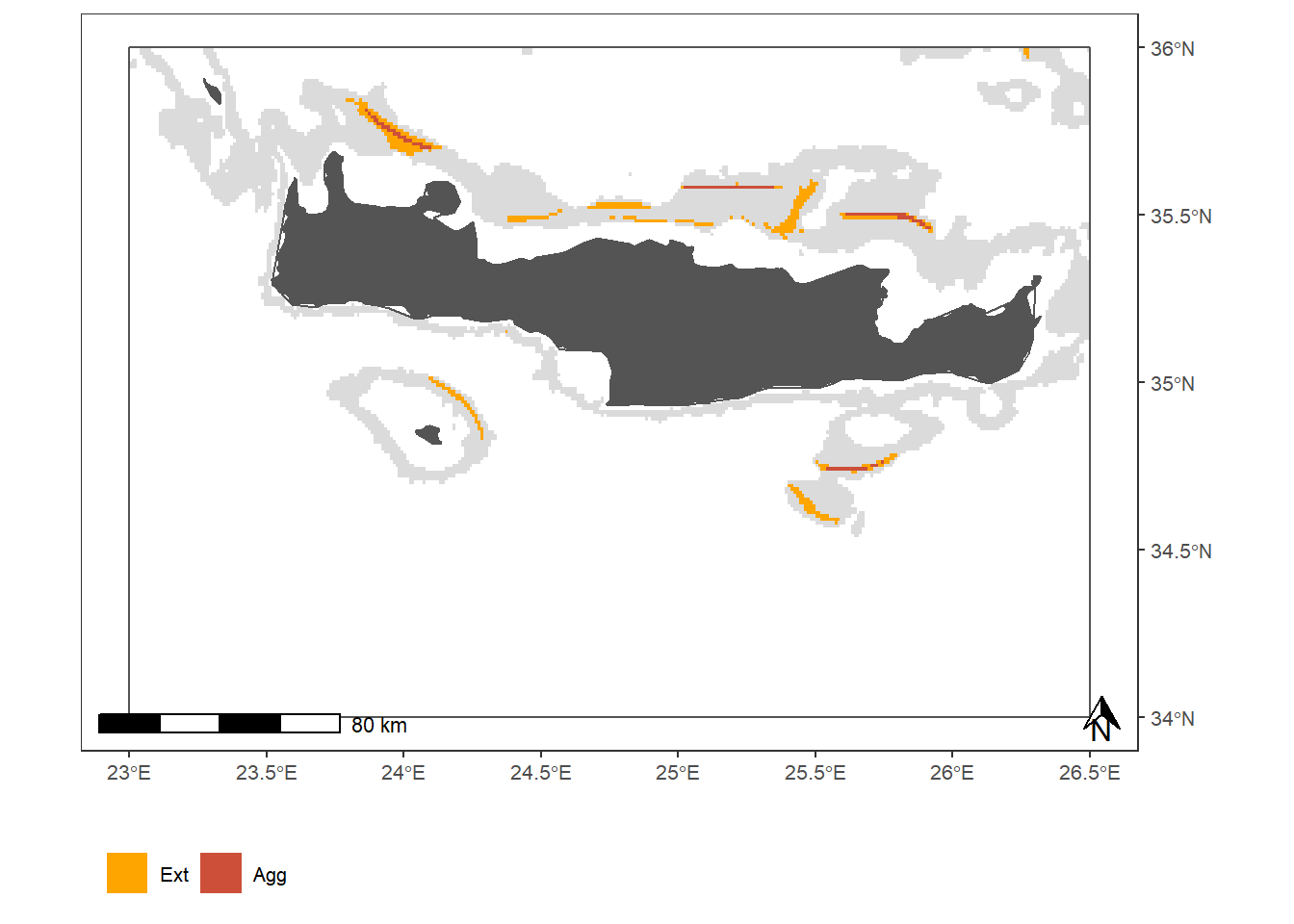
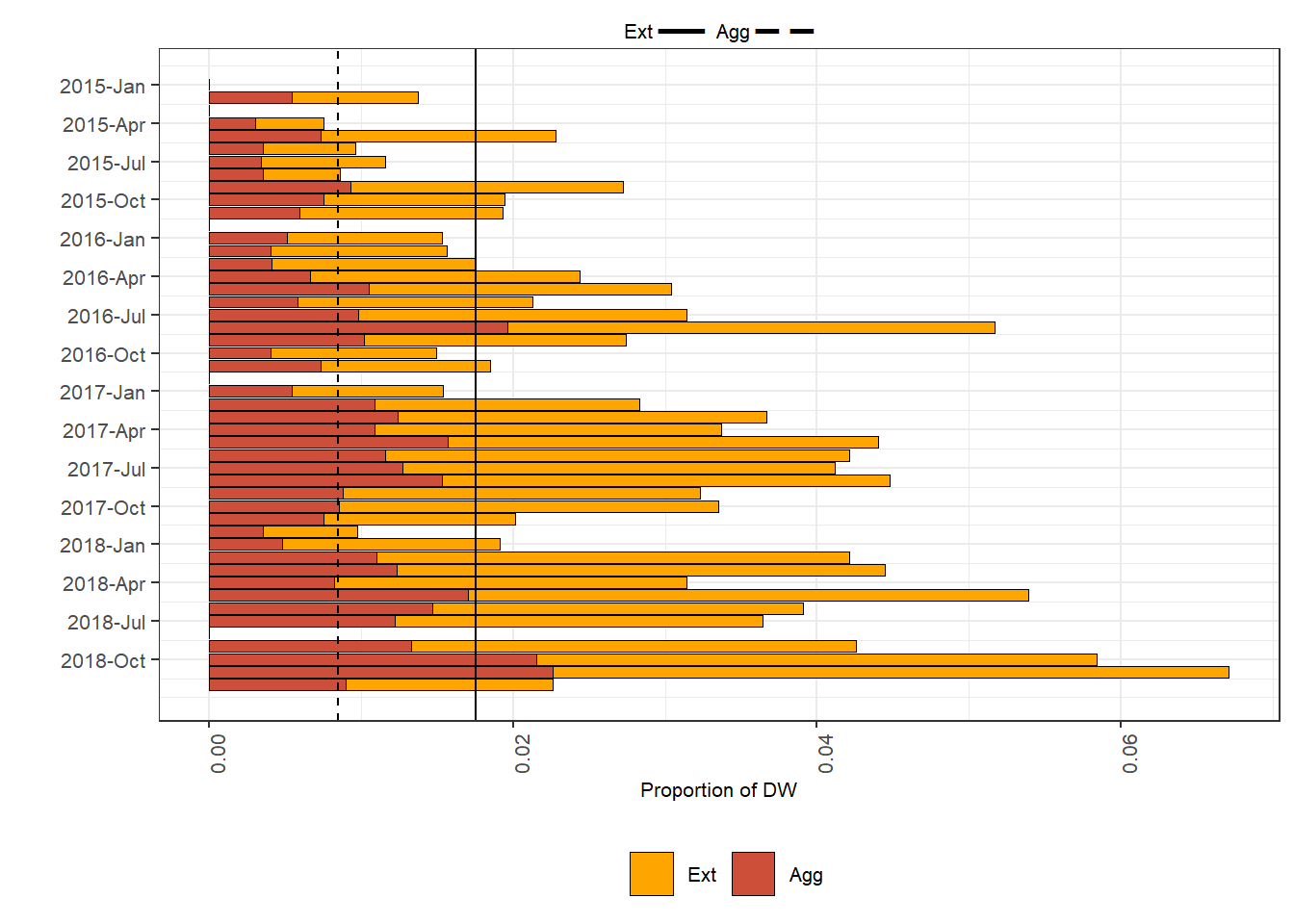
Indicators monthly time series
Step 1
The procedure is similar to what described in the Indicators map section, although the ind5 and ind6 functions here are applied on monthly basis.
First we define a temporal index, namely the month covering the period january 2015-december 2018
index=tibble(order=seq(1, 48, 1),
temporal_ref=seq(as.Date(paste("2015", "01", "01", sep = "-")),
as.Date(paste("2018", "12", "01", sep = "-")),"month"))| order | temporal_ref |
|---|---|
| 1 | 2015-01-01 |
| 2 | 2015-02-01 |
| 3 | 2015-03-01 |
| 4 | 2015-04-01 |
| 5 | 2015-05-01 |
| 6 | 2015-06-01 |
Then we build a for loop to iterate on each month contained in the temporal index. For each of the i timesteps, the data are filtered on the DW strata and the functions are applied. For each month we are interested in knowing the values assumed by the indicators, which in both cases are the number of cells falling in the indicator parameter. The ind_ts dataset serve to store all the monthly values, by adding the i value for each iteration over the time.
ind_ts=NULL
for (i in 1:nrow(index)){
# Subset data
idat=fishing_data[fishing_data$DW==1&
fishing_data$year==lubridate::year(index[i,]$temporal_ref)&
fishing_data$month==lubridate::month(index[i,]$temporal_ref),]
if(nrow(idat)==0){
next
}else{
i5=ind5(idat)
idat=aggregate(list('f_hours'=idat$f_hours), by=list('G_ID'=idat$G_ID), FUN=sum)
i6=ind6(idat)
ind_ts = rbind(ind_ts, data.frame(temporal_ref=index[i,]$temporal_ref, dcf5=length(i5),dcf6=nrow(i6)))
rm(i5,i6, idat)
}
}After having calculated the indicator monthly values, we standardize them over the GSA extension.
grid_cells=data.frame(table(grid$DW, grid$GSA))
ind_ts$ncell_rbs=grid_cells[grid_cells$Var1==1,]$Freq
ind_ts$dcf5_ratio = round(ind_ts$dcf5/ind_ts$ncell_rbs,4)
ind_ts$dcf6_ratio = round(ind_ts$dcf6/ind_ts$ncell_rbs, 4)
ind_ts$dcf_ratio = round((ind_ts$dcf6)/(ind_ts$dcf5),4)
ind_ts$temporal_ref=as.Date(ind_ts$temporal_ref)| temporal_ref | dcf5 | dcf6 | ncell_rbs | dcf5_ratio | dcf6_ratio | dcf_ratio |
|---|---|---|---|---|---|---|
| 2015-02-01 | 59 | 38 | 7076 | 0.0083 | 0.0054 | 0.6441 |
| 2015-04-01 | 32 | 21 | 7076 | 0.0045 | 0.0030 | 0.6562 |
| 2015-05-01 | 110 | 52 | 7076 | 0.0155 | 0.0073 | 0.4727 |
| 2015-06-01 | 43 | 25 | 7076 | 0.0061 | 0.0035 | 0.5814 |
| 2015-07-01 | 58 | 24 | 7076 | 0.0082 | 0.0034 | 0.4138 |
| 2015-08-01 | 36 | 25 | 7076 | 0.0051 | 0.0035 | 0.6944 |
Step 2
The monthly fishing hours and vessel statistics are calculated only for those months were fishing activity was actually observed. However we aim to consider in the plot also the months where no activity at all was observed. Therefore some formatting lines are needed.
ind_ts=merge(index, ind_ts, all.x = T)
ind_ts[is.na(ind_ts)]=0The dataset is reshaped to fit into the plot specifications
ind_ts=reshape2:::melt(ind_ts[,c('temporal_ref','order', 'dcf5_ratio', 'dcf6_ratio')], id.var=1:2)
ind_ts$variable=factor(ifelse(ind_ts$variable == "dcf5_ratio", "Ext", "Agg"), levels = c("Ext", "Agg"))| temporal_ref | order | variable | value |
|---|---|---|---|
| 2015-01-01 | 1 | Ext | 0.0000 |
| 2015-02-01 | 2 | Ext | 0.0083 |
| 2015-03-01 | 3 | Ext | 0.0000 |
| 2015-04-01 | 4 | Ext | 0.0045 |
| 2015-05-01 | 5 | Ext | 0.0155 |
| 2015-06-01 | 6 | Ext | 0.0061 |
Plot
Specifications for the plot
Plots are standardized between figures using the settings loaded at the beginning of the script. In addition, labeling can be optimized to fit the data of each area.
plot_breaks=seq(min(ind_ts$order),max(ind_ts$order),3)
plot_labels=paste(substr(seq(min(ind_ts$temporal_ref),max(ind_ts$temporal_ref),"quarters"),1,4),
month.abb[as.numeric(substr(seq(min(ind_ts$temporal_ref),max(ind_ts$temporal_ref),"quarters"),6,7))], sep="-")Plot it!
plot_indicators_ts = defo_ts +
geom_bar(data = ind_ts , aes(x=order, value, fill = variable), stat = "identity", colour = 1, na.rm = F, size=0.2) +
scale_fill_manual(values = c("orange", "tomato3")) +
scale_x_reverse(breaks=plot_breaks, labels=plot_labels)+
geom_hline( aes(yintercept = mean(ind_ts[is.na(ind_ts$value)==F & ind_ts$variable =="Ext", ]$value)))+
geom_hline( aes(yintercept = mean(ind_ts[is.na(ind_ts$value)==F & ind_ts$variable =="Agg", ]$value)), linetype=2)+
xlab("") +
ylab("Proportion of DW") +
guides(fill = guide_legend(title = "Indicator", size = 15), linetype=guide_legend())+
scale_y_continuous(sec.axis = sec_axis(~ . /50,breaks=c(1),labels="", name=expression(paste(""[""["Ext "]], bold("__"), ""[""[" Agg "]],bold("_ _") ))), name=expression(""[""["Proportion of DW"]]));plot_indicators_ts## Warning in min(x): no non-missing arguments to min; returning Inf## Warning in max(x): no non-missing arguments to max; returning -Inf